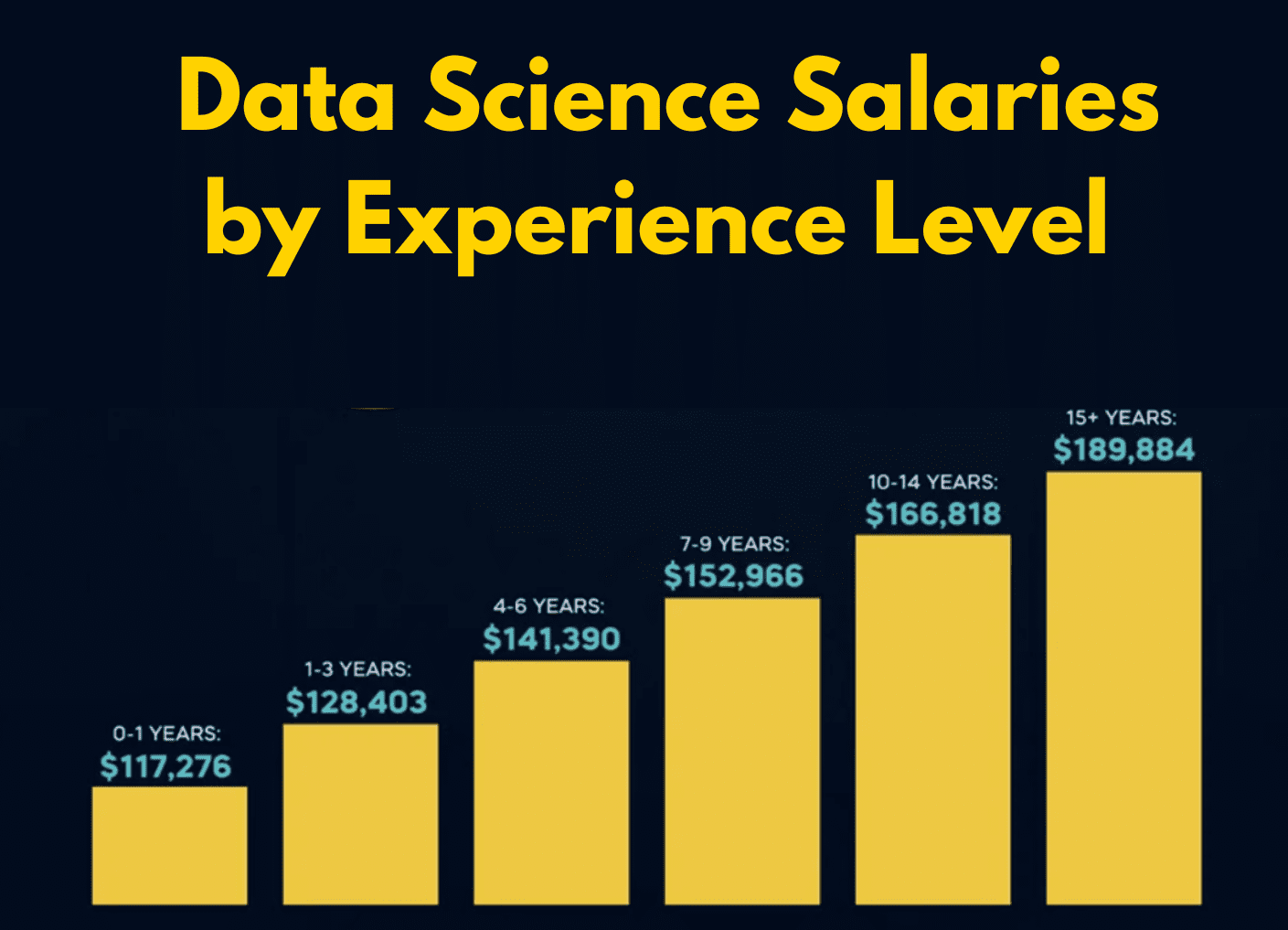
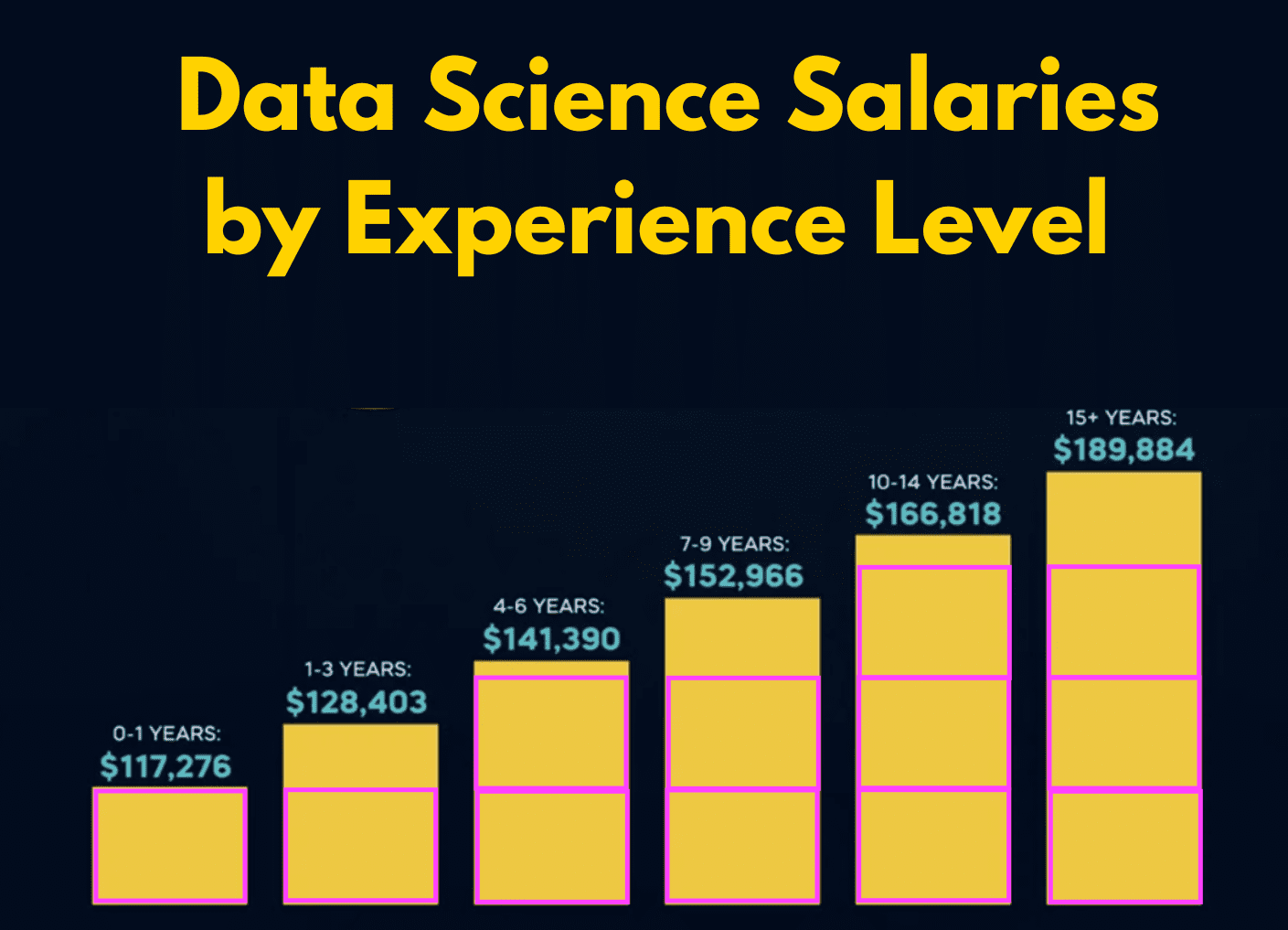
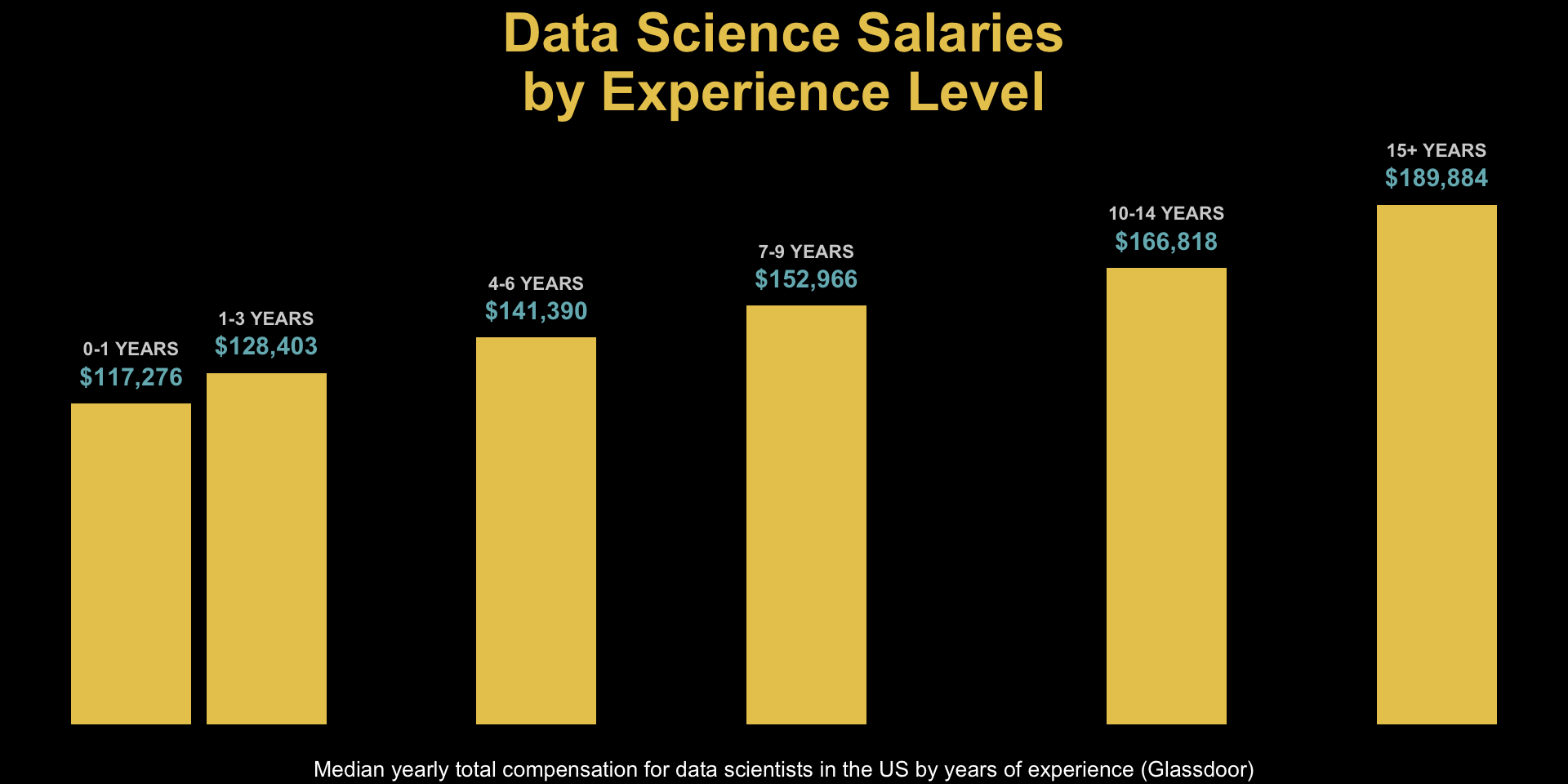
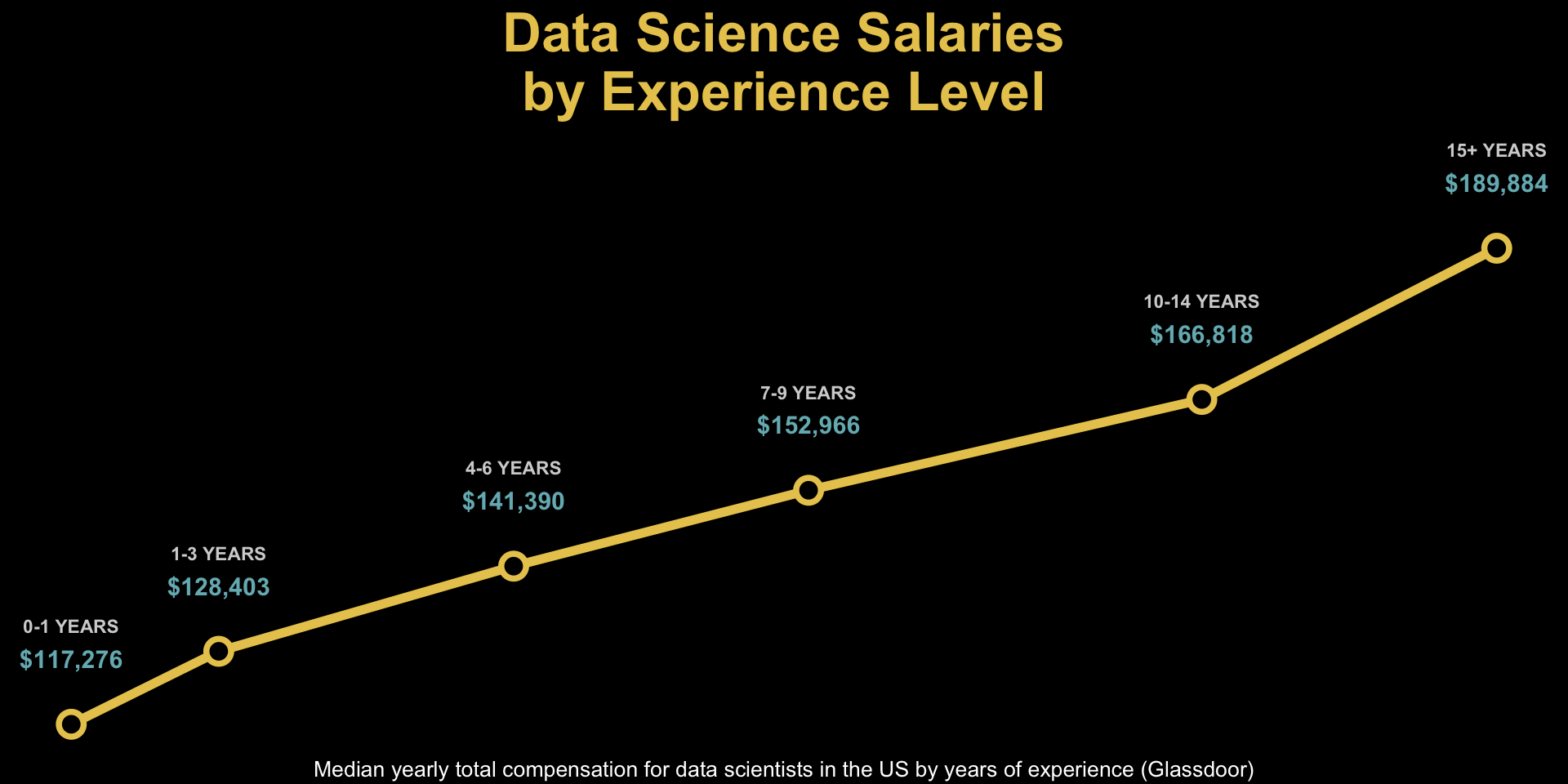
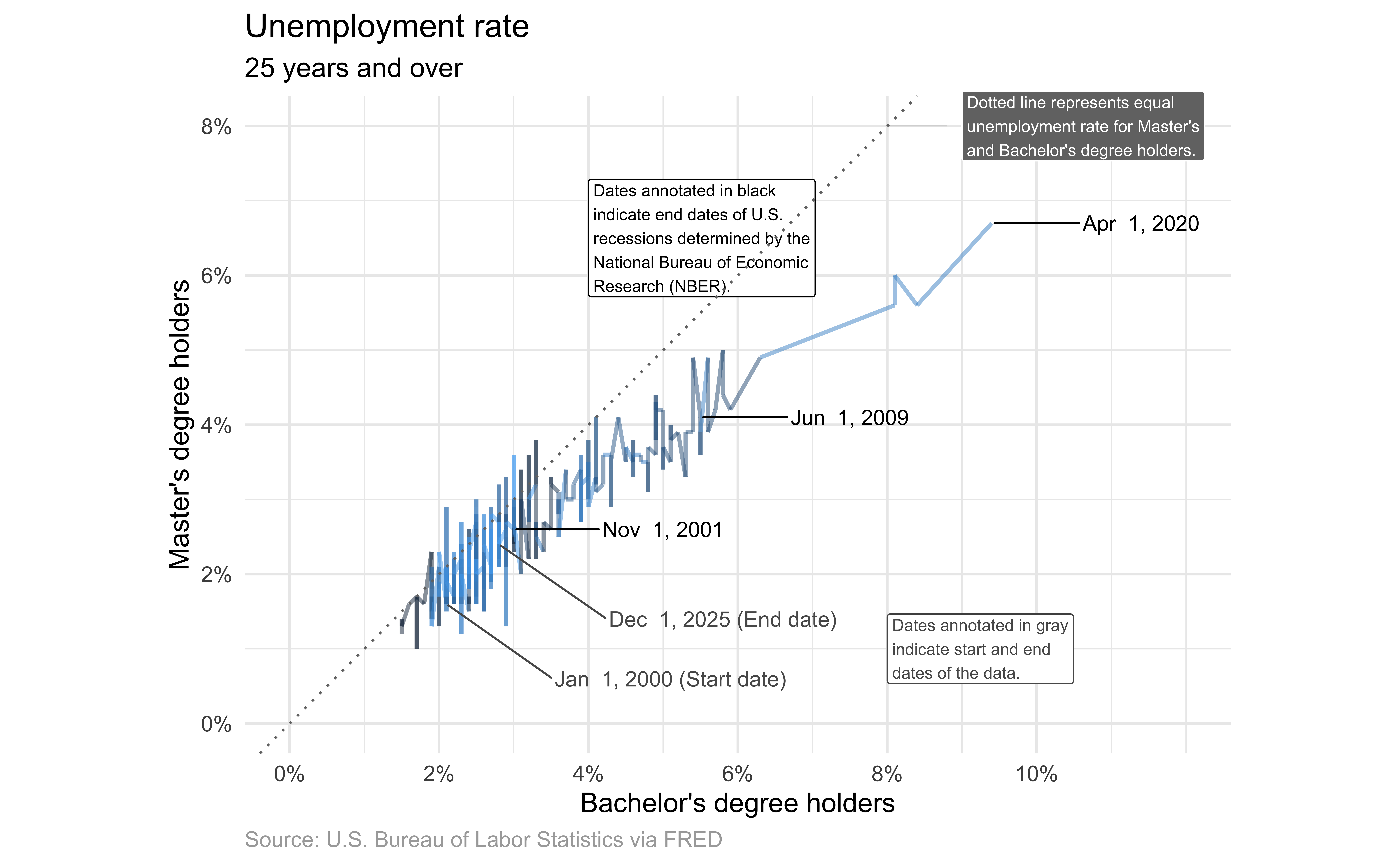
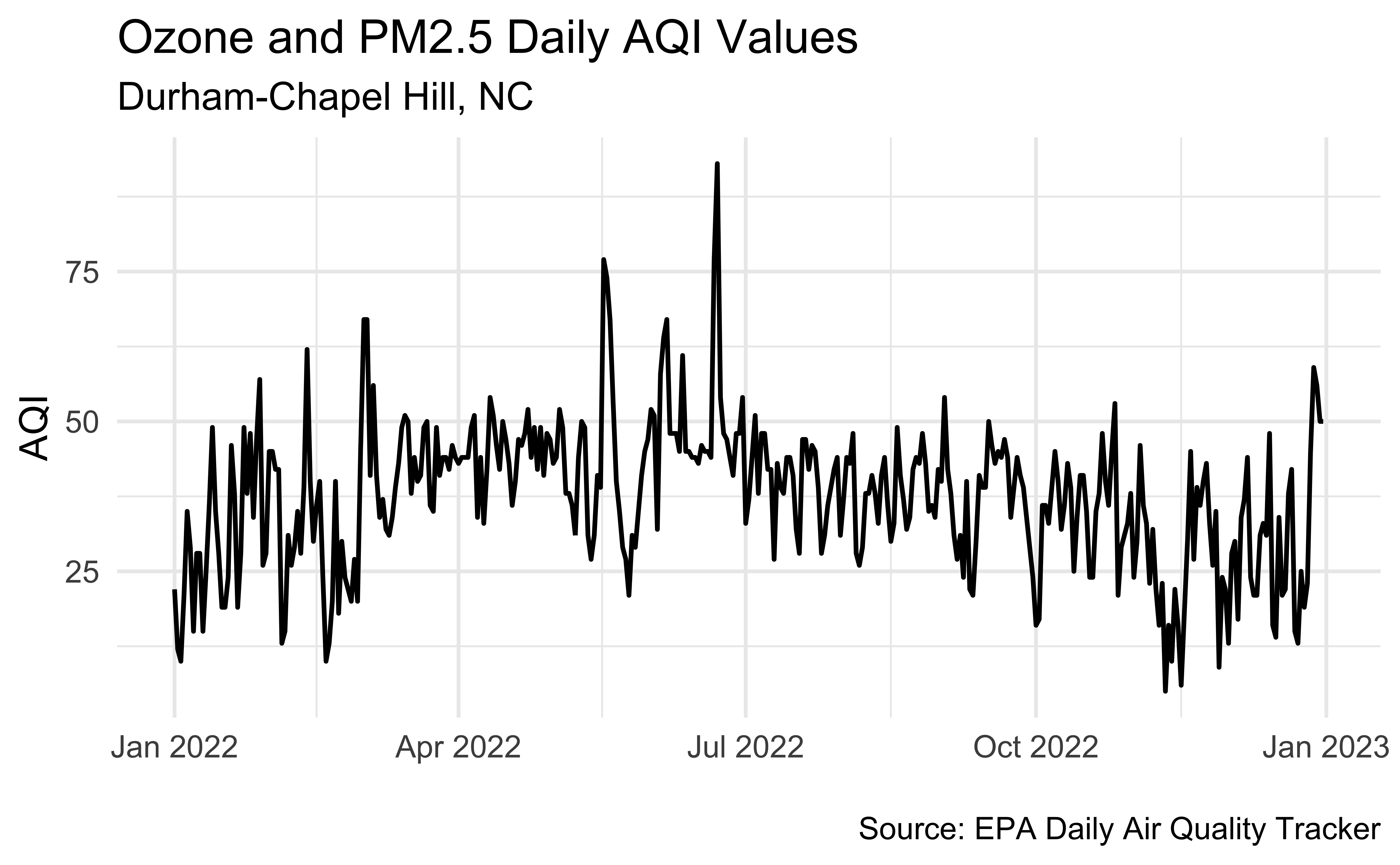
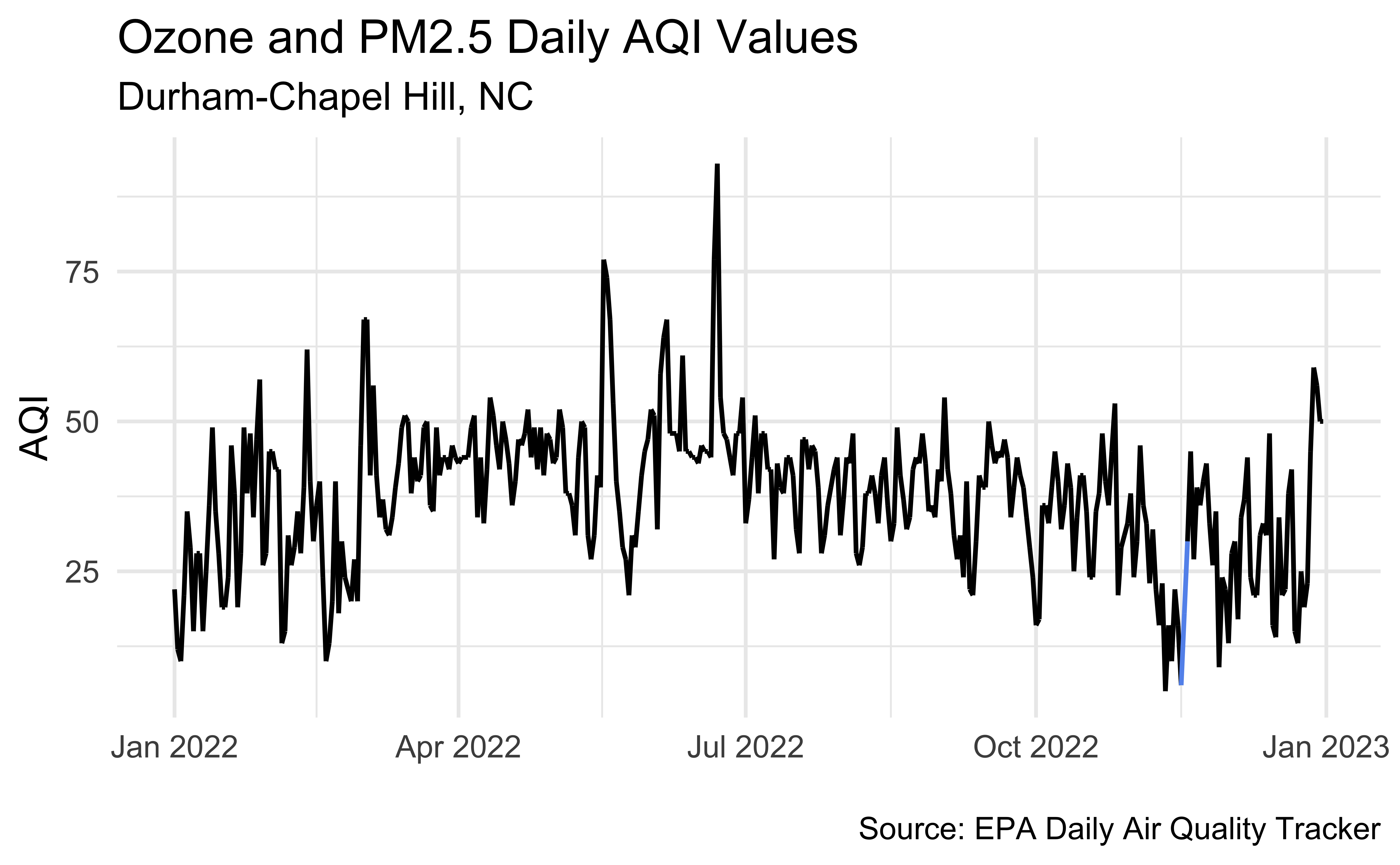
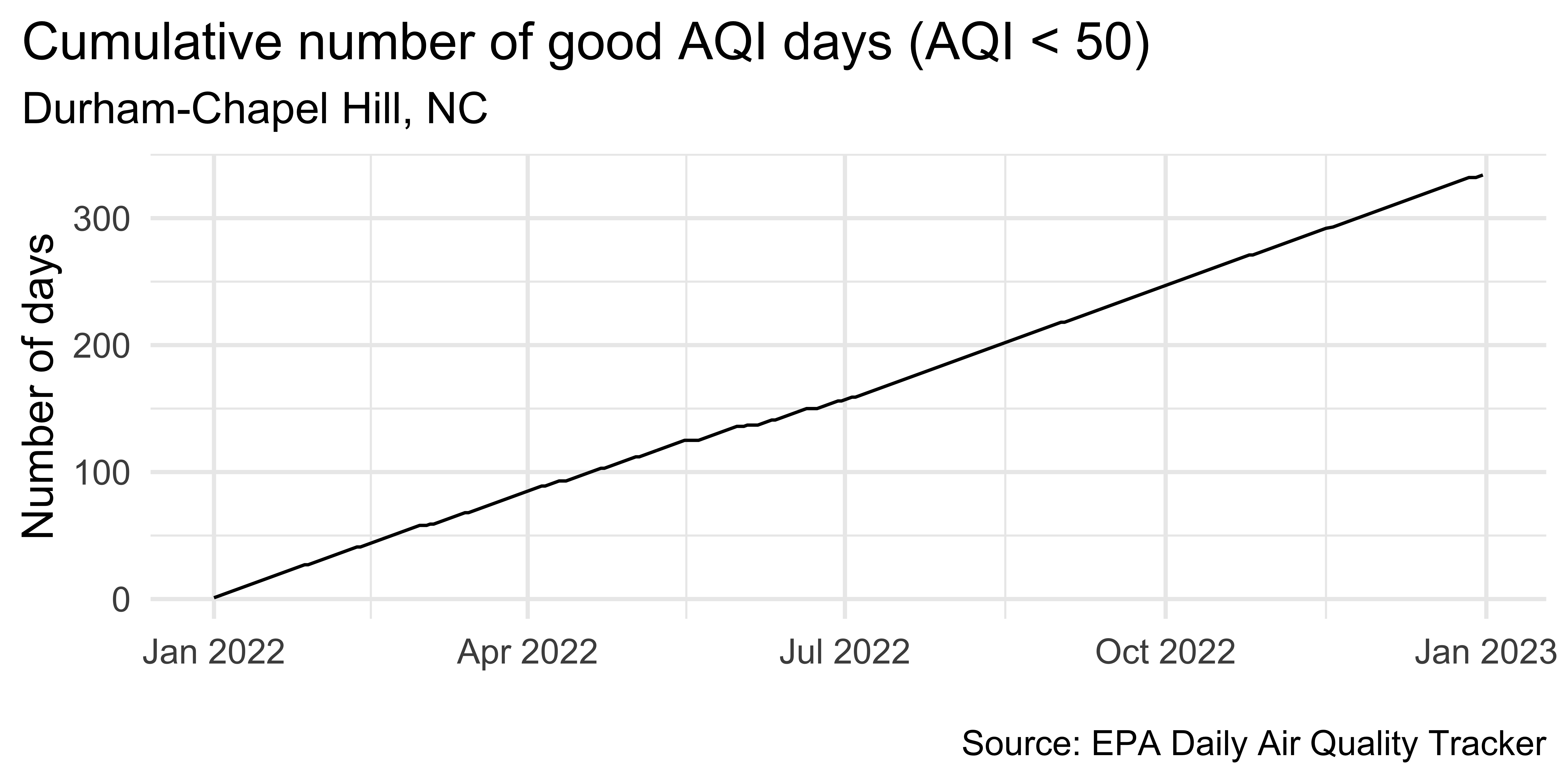
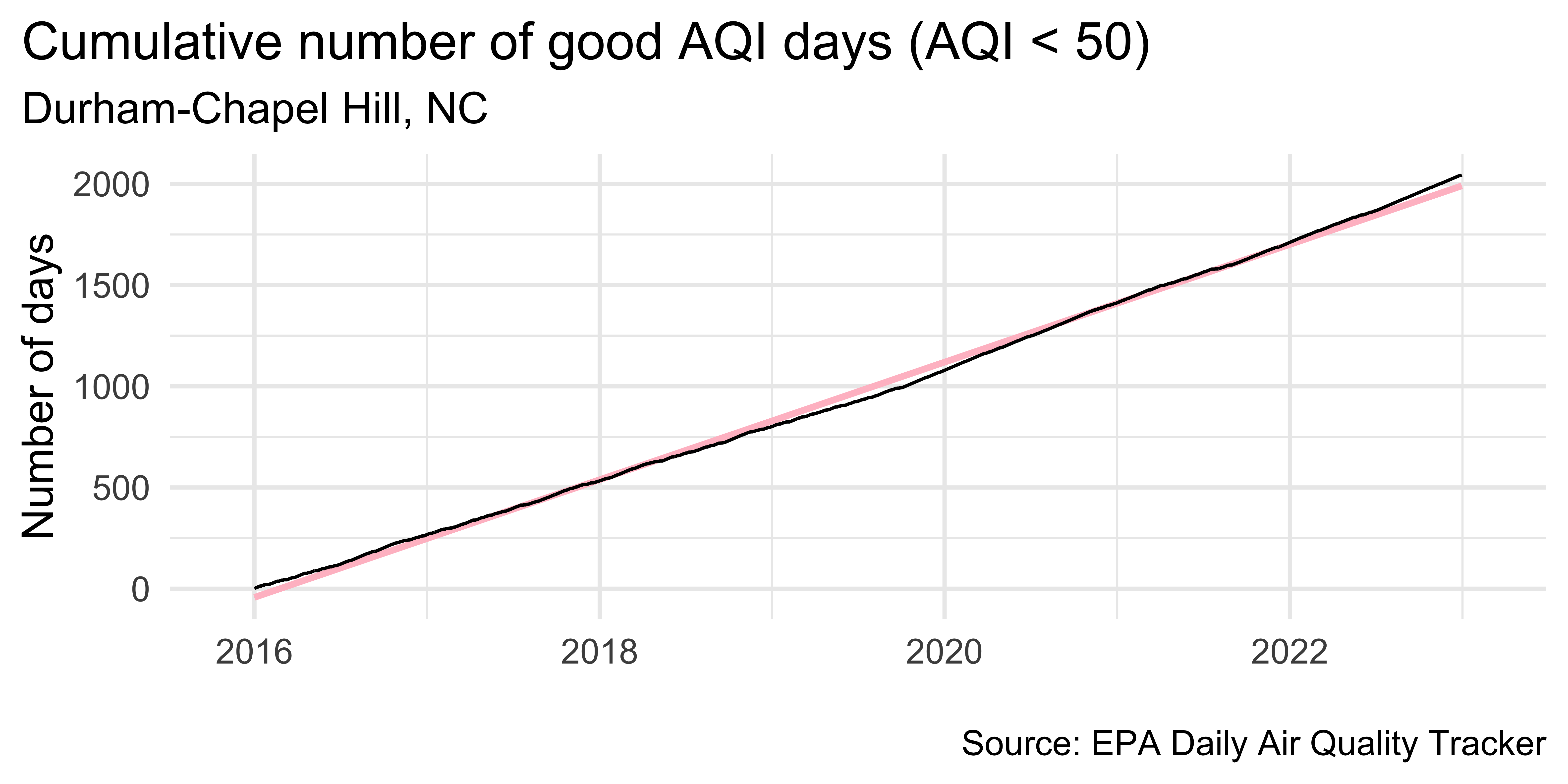
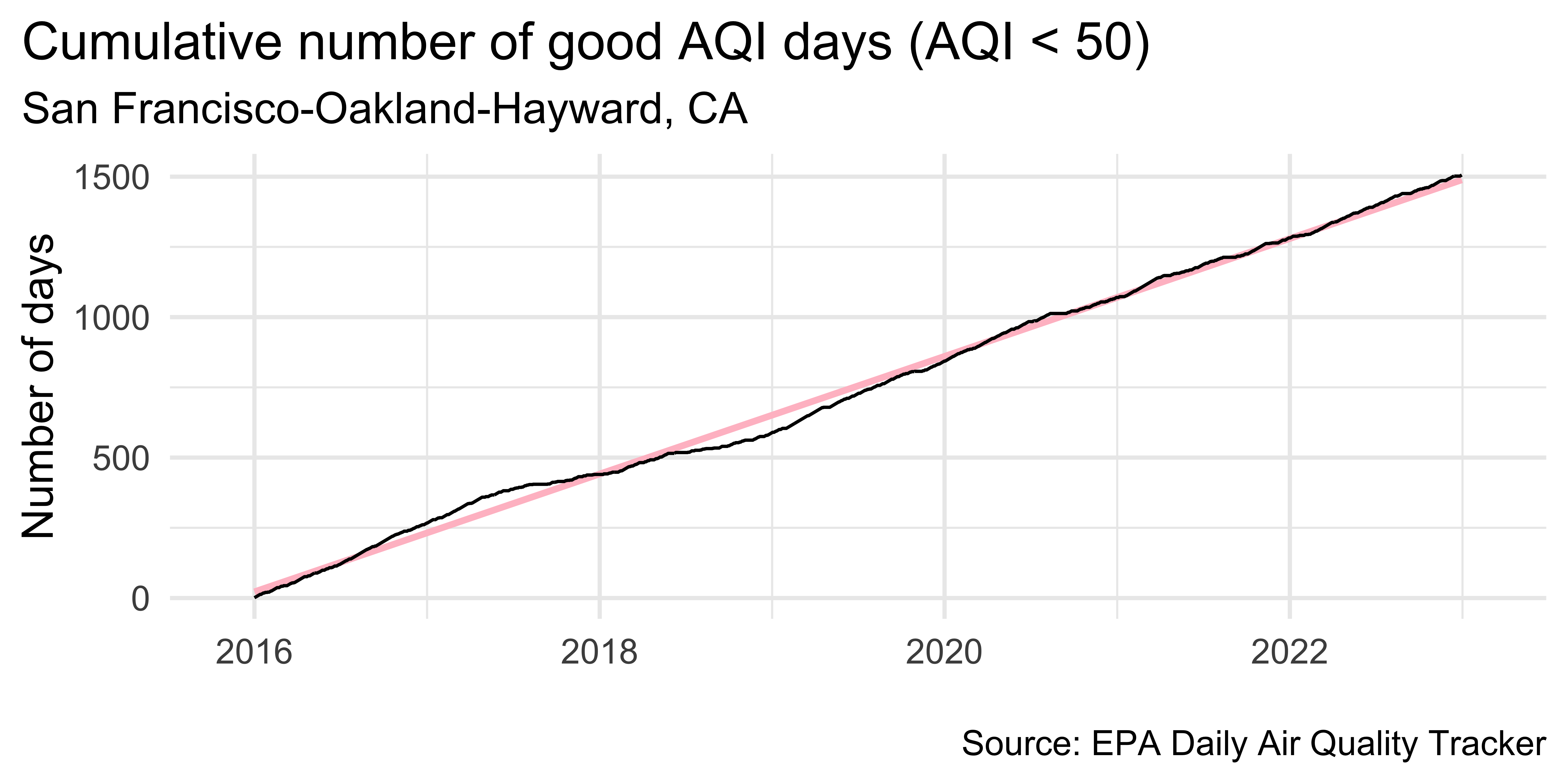
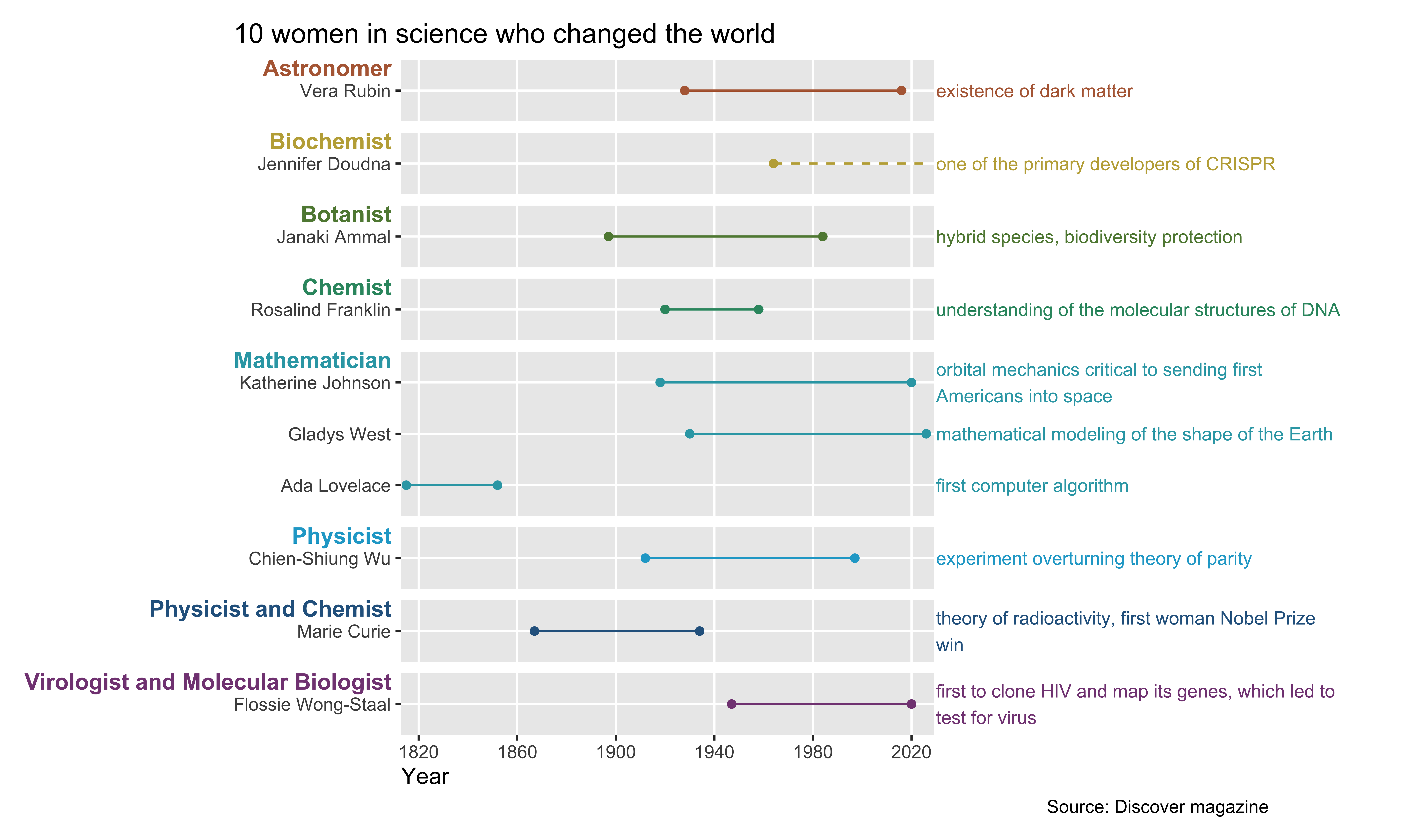
unemp_bachelor_master_wider <- unemp |>
filter(degree %in% c("Bachelor's", "Master's")) |>
pivot_wider(names_from = degree, values_from = unemp_rate)
recession_end_dates <- c(
ymd("2001-11-01"),
ymd("2009-06-01"),
ymd("2020-04-01")
)
endpoints <- c(
ymd("2000-01-01"),
ymd("2025-12-01")
)
gray_dark_hex <- "#595959"
gray_light_hex <- "#737373"
ggplot(unemp_bachelor_master_wider, aes(x = `Bachelor's`, y = `Master's`)) +
geom_line(
aes(color = date),
linewidth = 1,
alpha = 0.5,
show.legend = FALSE
) +
scale_x_continuous(
labels = label_percent(scale = 1),
breaks = seq(0, 10, 2),
limit = c(0, 12)
) +
scale_y_continuous(
labels = label_percent(scale = 1),
breaks = seq(0, 10, 2),
limit = c(0, 8)
) +
geom_text_repel(
data = unemp_bachelor_master_wider |>
filter(date %in% recession_end_dates),
aes(label = as.character(format(date, "%b %e, %Y"))),
position = ggpp::position_nudge_keep(x = 2)
) +
# recessions
geom_text_repel(
data = unemp_bachelor_master_wider |>
filter(date %in% endpoints),
aes(
label = paste(
as.character(format(date, "%b %e, %Y")),
c("(Start date)", "(End date)")
)
),
position = ggpp::position_nudge_keep(x = 3, y = -1),
color = gray_dark_hex
) +
annotate(
geom = "label",
x = 4,
y = 6.5,
label = str_wrap(
"Dates annotated in black indicate end dates of U.S. recessions determined by the National Bureau of Economic Research (NBER).",
30
),
size = 3,
hjust = 0,
fill = "white"
) +
# endpoints
annotate(
geom = "label",
x = 8,
y = 1,
label = str_wrap(
"Dates annotated in gray indicate start and end dates of the data.",
25
),
size = 3,
hjust = 0,
fill = "white",
color = gray_dark_hex
) +
# diagonal
geom_abline(
slope = 1,
intercept = 0,
linetype = "dotted",
color = gray_light_hex
) +
annotate(
geom = "segment",
x = 8,
xend = 8.8,
y = 8,
yend = 8,
linewidth = 0.2,
color = gray_light_hex
) +
annotate(
geom = "label",
x = 9,
y = 8,
label = str_wrap(
"Dotted line represents equal unemployment rate for Master's and Bachelor's degree holders.",
30
),
size = 3,
hjust = 0,
fill = gray_light_hex,
color = "white"
) +
labs(
title = "Unemployment rate",
subtitle = "25 years and over",
x = "Bachelor's degree holders",
y = "Master's degree holders",
caption = "Source: U.S. Bureau of Labor Statistics via FRED"
) +
theme(plot.caption = element_text(color = "darkgray", hjust = 0)) +
coord_equal(clip = "off")